Resources
Model Diagnosis
HYPERPLAne OPtimization for attribute boundary
We are learning the behavior of the diffusion latent space and exploring both the h-space (bottleneck of U-Net) and e-space (image space) for attribute manipulation. We’ve tried to find the hyperplane that corresponds to a given attribute in h-space and/or e-space. Below are some generated images as we traverse the normal vector to the discovered attribute hyperplane.

As you see above, the attribute manipulation feels limited via this method of hyperplane traversal. One key challenge is that, since diffusion models have many steps, we can make edits to the latent vector in many ways: at one step only, at each step, same edit at each step, different edit per step, etc. My feeling is that we have not found the most optimal way to manipulate the diffusion latent vector for semantic edits.
Implicit function edit of diffusion h-space and/or e-space
Following the work above, training an implicit function would enable us to make step-dependent edits to the diffusion latent vector. The implicit function takes as input a step s and the h-space vector to produce delta_h, which is then added to the h-space vector in the bottleneck layer of U-Net. We’ve experimented with extending this idea to latent diffusion models, but the results are not convincing.
LLM-Aided attribute discovery

In this line of work, we established a proof-of-concept by:
- First prompting GPT-4 to get relevant attributes for a given domain
- Query text2image to generate 100s of examples for each attribute from 1.
- Run a target model on these images to get a coarse sensitivity score
The next steps would be to run a deeper sensitivity analysis using one of the frameworks HYPERPLANE OPTIMIZATION or IMPLICIT FUNCTION EDIT.
Data Augmentation
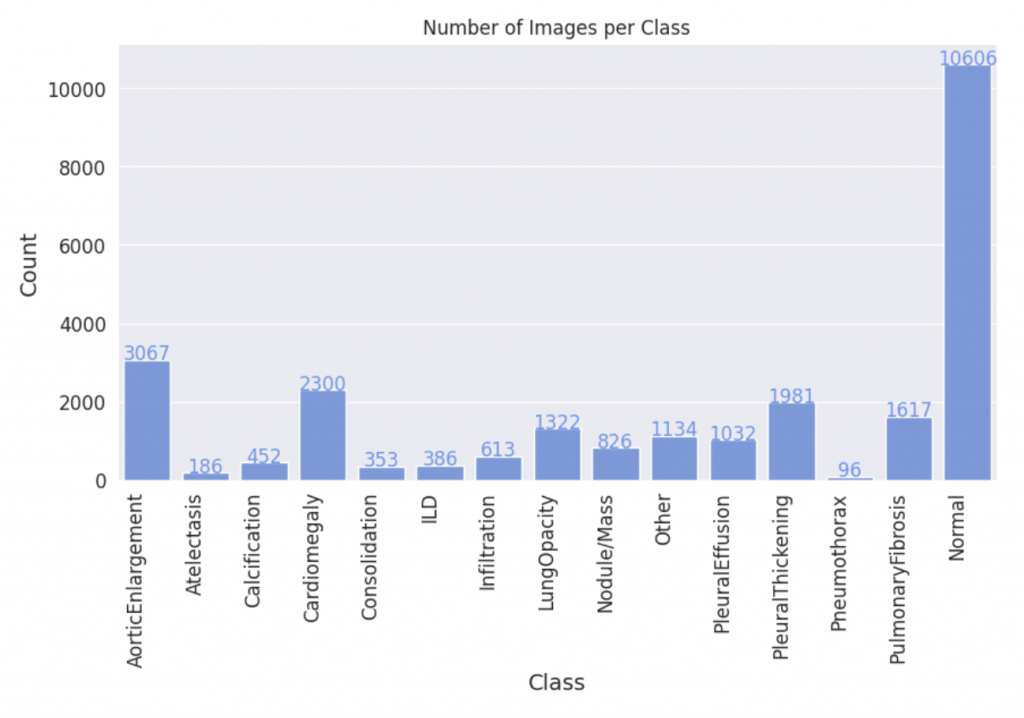
Given the scarcity of data in medical AI, we have identified the medical domain as a primary area of focus for our research. Specifically, we aim to expand the widely-used VinDR-CXR dataset, which includes 15,000 accurately labeled chest X-ray images across 14 disease categories. However, the imbalanced distribution of images across these categories presents a challenge, as some disease classes have significantly fewer examples than others. To address this, we propose generating synthetic images for these underrepresented classes, and assessing the quality of these generated images through a comprehensive evaluation of downstream classification models trained on both synthetic and real images.
We fine-tuned the Stable Diffusion 1.4v model with the DreamBooth approach, utilizing a small set of 3-5 images from each class within the target domain. Qualitatively, the generated images exhibit a noticeable improvement in resemblance to real images from the target domain.
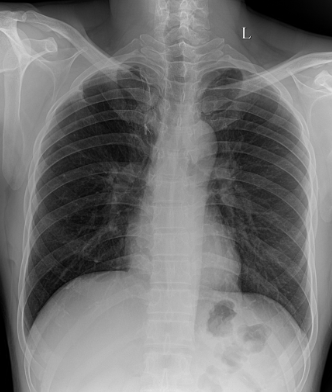
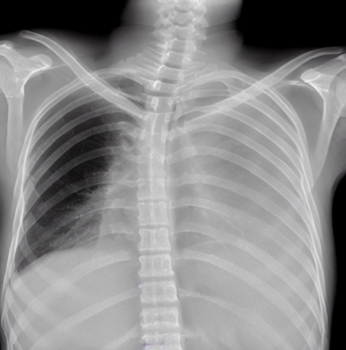
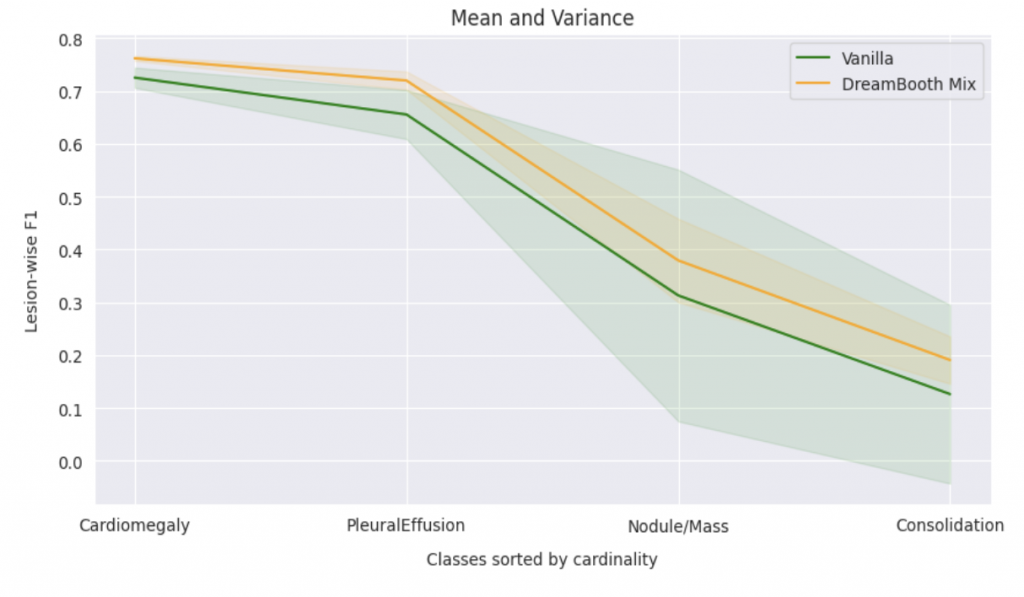
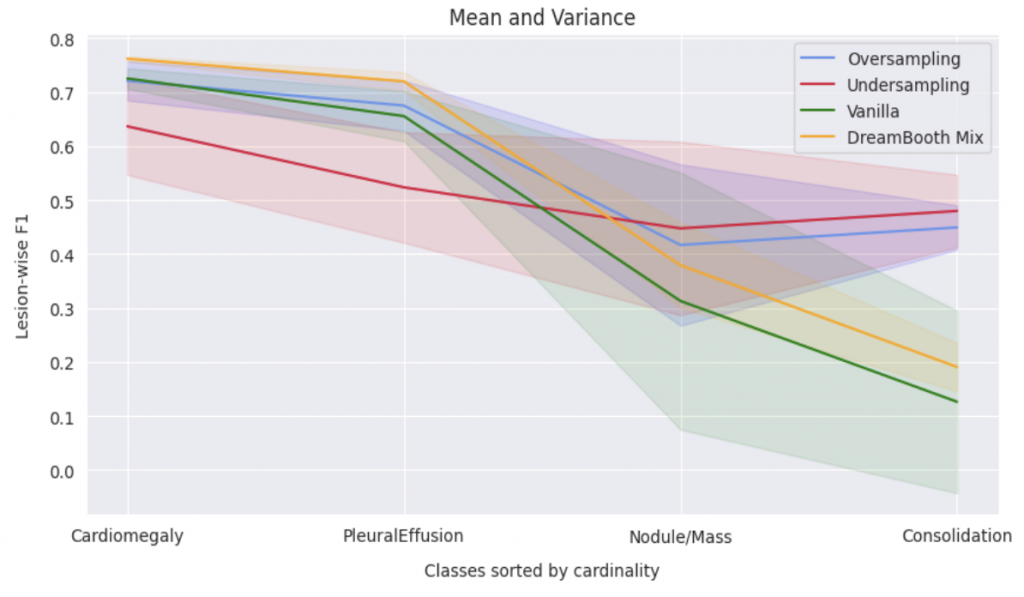
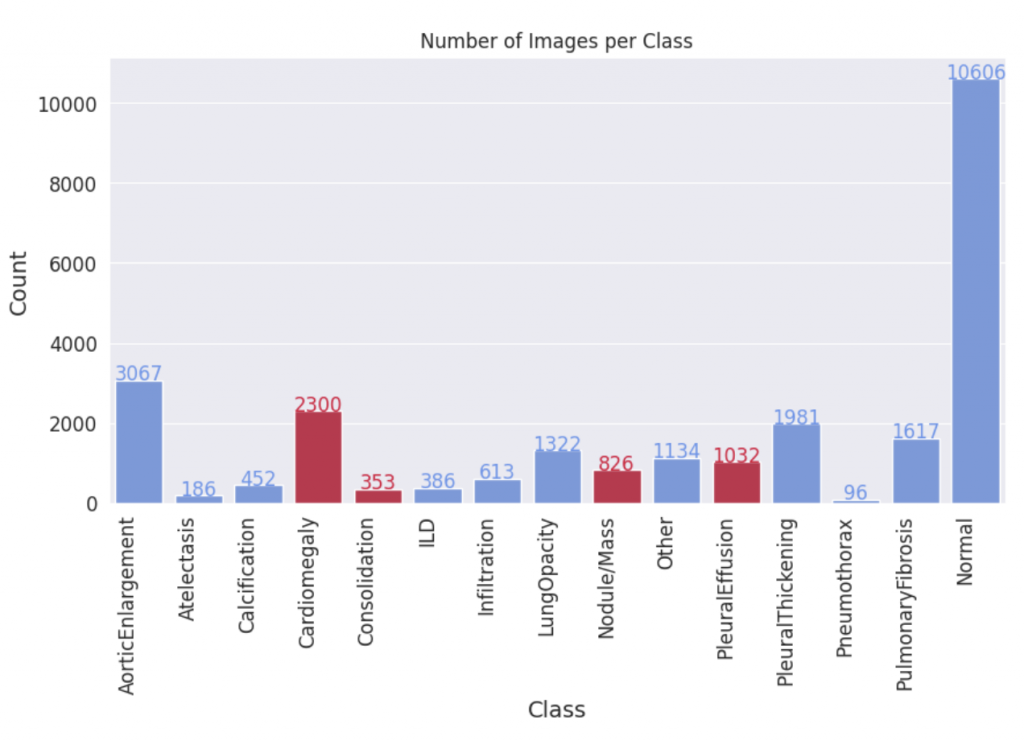
In order to evaluate the quantitative impact of augmenting the dataset with synthetic images, we employed ResNet-50, pre-trained on ImageNet, to train a multi-label classification model on a four-class task. The synthetic images were generated to ensure even distribution of images for each class in the training dataset. We compared this method against three baseline approaches, including vanilla training with unbalanced classes, as well as under- and over-sampling methods with balanced classes. The quality of the generated images was assessed by measuring the performance of downstream classification models trained on these datasets.
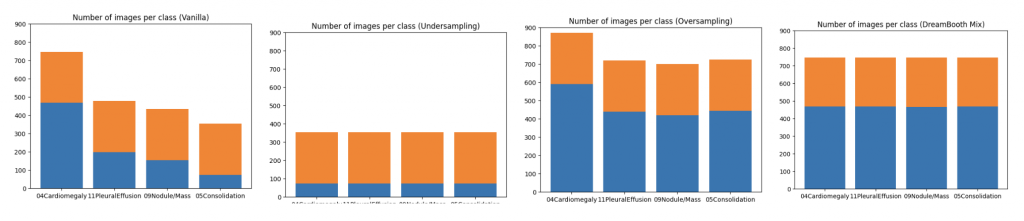
When compared to vanilla method, the DreamBooth Mix (synthetic image) achieves consistent improvements in all 4 lesions. It was also found to effectively address domain gaps in the majority classes, while undersampling led to a degradation in performance for these classes. Oversampling, on the other hand, resulted in no domain gaps between the training and added data, but was lacking in diversity. The approach will be further evaluated on more extreme cases, where only 5-20 real images are available for minority classes.