Evaluation Datasets
We test the various restoration methods with several different test datasets. The EAD2019 dataset (~600 images) provides us with artifact segmentation labels, so we use this dataset to evaluate our models’ abilities to restore a variety of artifacts. The ENDO4IE dataset (~300 images) was split up into two datasets. One dataset was created by applying continuous random masks on clean images, in order to evaluate how well a model recovered color. The other dataset was created by hand-picking endoscopic images with lots of geometric structure, and applying pseudo-random masks on specific regions that contained lots of structure. Both datasets contain images that are from the GI tract and colonoscopy procedures.
Results On EAD2019 Test Set
Below are quantitative and qualitative results on the EAD2019 test dataset. We use single image metrics, including PIQE and SNR, since this dataset does not provide the corresponding “clean” version of the input image.


Results on ENDO4IE Color Recovery Test Set
Below are quantitative and qualitative results on the ENDO4IE color recovery test dataset. In order to evaluate the color accuracy, we use PSNR as the image quality metric. This will tell us how closely the restoration model was at inpainting the original pixel color.

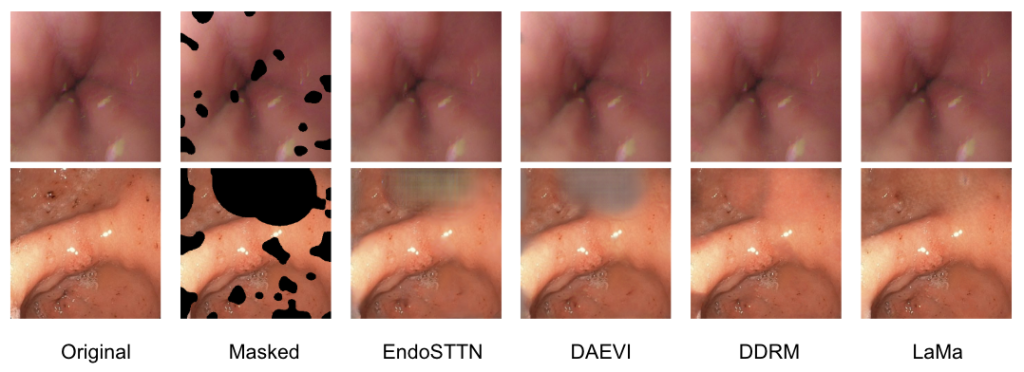
Results on ENDO4IE Structure Recovery Test Set
Below are quantitative and qualitative results on the ENDO4IE structure recovery test dataset. In order to evaluate the color accuracy, we use SSIM as the image quality metric. This metric evaluates structure that human perception cares about, including brightness, contrast, and texture.


Analysis
From the results above, we select LaMa as our final model in our endoscopic image restoration pipeline. We can see that the temporal GAN methods are unable to recover large artifact areas and produce non-meaningful results. Diffusion methods aren’t able to accurately recover realistic structure, as DDRM simply splats a flat, uniform blob on the masked regions. However, we can see that LaMa performs well at recovering large masks and geometric structure.
YOLOv11 Segmentation Finetuning
We finetune the YOLOv11 segmentation model with EAD data. The images in the EAD dataset have variable sizes, and YOLOv11 can only operate on image sizes that are to the power of 2. Therefore, we created several 64×64 crops of each image to prepare our dataset. We train the model for 200 epochs. The qualitative results are shown below.